Archives
- 2025-12
- 2025-11
- 2025-10
- 2025-09
- 2025-03
- 2025-02
- 2025-01
- 2024-12
- 2024-11
- 2024-10
- 2024-09
- 2024-08
- 2024-07
- 2024-06
- 2024-05
- 2024-04
- 2024-03
- 2024-02
- 2024-01
- 2023-12
- 2023-11
- 2023-10
- 2023-09
- 2023-08
- 2023-07
- 2023-06
- 2023-05
- 2023-04
- 2023-03
- 2023-02
- 2023-01
- 2022-12
- 2022-11
- 2022-10
- 2022-09
- 2022-08
- 2022-07
- 2022-06
- 2022-05
- 2022-04
- 2022-03
- 2022-02
- 2022-01
- 2021-12
- 2021-11
- 2021-10
- 2021-09
- 2021-08
- 2021-07
- 2021-06
- 2021-05
- 2021-04
- 2021-03
- 2021-02
- 2021-01
- 2020-12
- 2020-11
- 2020-10
- 2020-09
- 2020-08
- 2020-07
- 2020-06
- 2020-05
- 2020-04
- 2020-03
- 2020-02
- 2020-01
- 2019-12
- 2019-11
- 2019-10
- 2019-09
- 2019-08
- 2019-07
- 2019-06
- 2019-05
- 2019-04
- 2018-11
- 2018-10
- 2018-07
-
Several main strategies for targeting E s described in the
2020-07-14
Several main strategies for targeting E3s described in the literature include: in vitro screening using functional assays [86]; computer programs for predicting potential druggable pockets, including those at protein-protein interfaces, and subsequent docking-based in silico ligand screening [87]; f
-
br Inhibition of DHODH The final products a n were
2020-07-14
Inhibition of DHODH The final products 7a–n were assessed for their DHODH inhibitory activity on rat liver mitochondrial/microsomal membranes. A procedure adapted from the literature was employed (see Experimental), in which oxidation of DHO to ORO is monitored by following the concomitant reduct
-
br Experimental section br Notes br PDB ID codes
2020-07-14
Experimental section Notes PDB ID codes The atomic coordinates and structure factors of hDHODH in complex with compounds 4 (PDB id: 5MVC), 5 (PDB id: 5MVD) and 6 (PDB id: 5MUT) have been deposited in the RCSB Protein Data Bank. Acknowledgements This research was financially supported b
-
To compare glucose metabolism via oxidative phosphorylation
2020-07-14
To compare glucose metabolism via oxidative phosphorylation to that via glycolysis, a bioenergetics plot was constructed (Fig. 3A). In most cells, a decrease in one bioenergetics pathway is compensated by an increase in the other. However, following NAT1 deletion, there was a decrease in both oxidat
-
Six substrate recognition sites SRS
2020-07-14
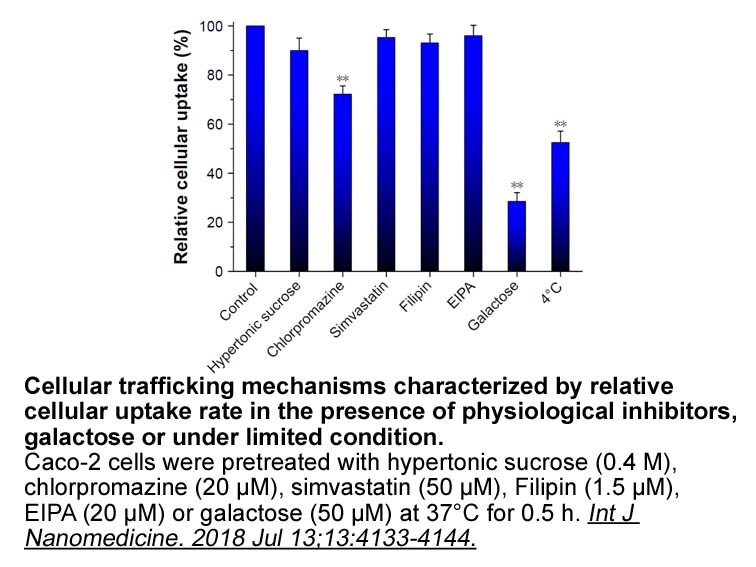
Six substrate recognition sites (SRS1-SRS6) have been identified at positions 102-123, 204-209, 238-244, 301-319, 374-382 and 483-492 (Table 2, underlined residues) based on Gotoh\'s proposal [30]. CYP3A163 displays the similar characteristic long sequences between helices F and G presenting two add
-
DDX belongs to the DEAD box family of proteins ATP
2020-07-13
DDX3 belongs to the DEAD-box family of proteins, ATP-dependent RNA helicases characterized by the presence of a highly conserved helicase core domain [13]. This catalytic core is composed of two RecA-like domains containing motifs involved in ATP binding/hydrolysis, RNA binding and helicase activity
-
itk inhibitor Several RNA BPs like the ELAV protein family
2020-07-13

Several RNA-BPs, like the ELAV protein family member HuR, tristetraprolin (TTP) or the KH-type splicing regulatory protein (KSRP) have been shown to interact with these AREs and thereby lead to stabilization or destabilization of the mRNA [5]. Control of nucleocytoplasmic mRNA export is also very i
-
br Clinical potential of ET receptor biased ligands Is
2020-07-13
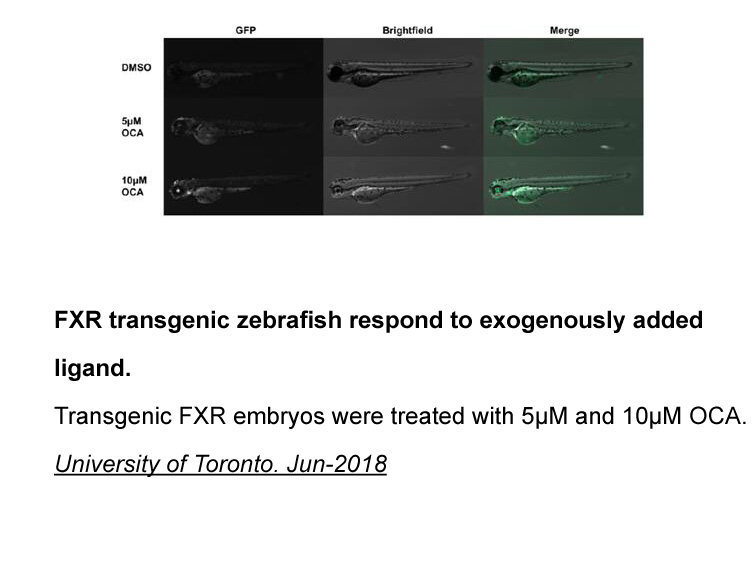
Clinical potential of ET receptor biased ligands Is there a clinical need for ET receptor biased ligands? Evidence is strongest from research in epithelial ovarian cancer demonstrating that ET-1 stimulated ETA-mediated β-arrestin signalling leads to activation of the oncogenic mediator NF-κB [21]
-
The UPR is composed of three different
2020-07-13

The UPR is composed of three different pathways that fall under the control of three respective ER transmembrane proteins: PERK, IRE1α (inositol-requiring enzyme 1α) and ATF6 (activating transcription factor 6). As a starting signal for the UPR, misfolded proteins induce the release of GRP78 from th
-
br Experimental Procedures br Acknowledgments The authors wi
2020-07-13
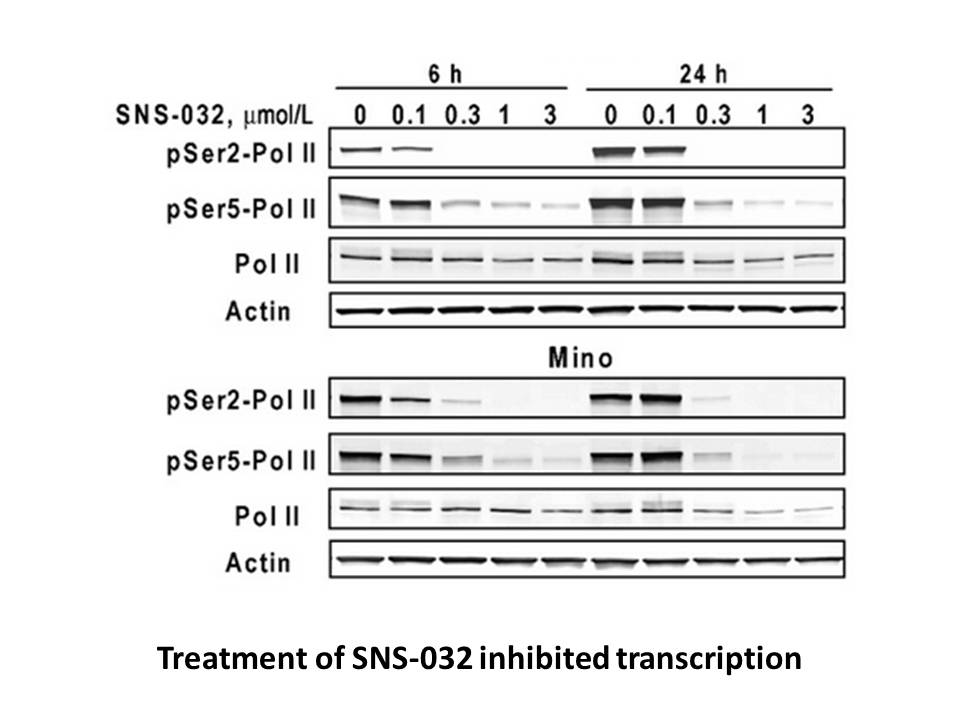
Experimental Procedures Acknowledgments The authors wish to thank P. Costet (University of Bordeaux) and Véronique Guyonnet-Duperat (FR TransBioMed, Plateforme de Vectorologie, University of Bordeaux) and Prof. Nils-Göran Larsson and Dr. Bettina Bertalan (Max Planck Institute for Biology of Ag
-
In conclusion we have discovered two new derivatives and tha
2020-07-13
In conclusion, we have discovered two new derivatives ( and ) that are potent inhibitors of DHODH. H and C NMR spectroscopic data revealed that these compounds undergo ready isomerisation at room temperature in -DMSO, but the docking studies indicate that there is neither conformation nor configurat
-
As shown in Figs a and b with
2020-07-13
As shown in Figs. 3(a) and (b), with the SO effect, the absolute values of the VDEs change by at most 0.19eV. Table 1 also shows that the observed peak splitting energies in the Sm-Ho complexes were reproduced even without the SO effect. Considering its significant importance in the electronic struc
-
p-Cresyl sulfate sale A topological property P is
2020-07-13
A topological property P is said to be cut point hereditary [3] if whenever a space X has property P, then, for every there exists some separation of such that each of and has property P. A space X is called connected ordered topological space (COTS) [5] if it is connected and has the property
-
Introduction Human cytomegalovirus CMV is a
2020-07-13
Introduction Human cytomegalovirus (CMV) is a herpesvirus which has a ubiquitous and worldwide distribution, and is the most frequent cause of congenital infection (Kenneson and Cannon, 2007). The prevalence is around 0.5–1% of all live births and is the leading cause of sensorineural hearing loss
-
DC_AC50 The action of pt PGE
2020-07-11
The action of 17-pt-PGE2 as an EP1 receptor agonist [34] was demonstrated in several experimental settings, including cancer, neurons, vascular system, kidney and was confirmed by application of EP1 receptor selective antagonists [35], [36], [37], [38], [39], [40], [41]. Receptor binding studies in
15780 records 793/1052 page Previous Next First page 上5页 791792793794795 下5页 Last page